Molecular interaction networks I & II
Molecular interaction networks
Credits:
Manuela Garretón I.
Teacher supervisor:
Rodrigo Gutiérrez
José Miguel Tagle
Pablo Hermansen
Bioligist Research:
Felipe Aceituno
Completion: 2007
Research financed by Fondecyt* nº 1060457
These images correspond to my design thesis project, which was developed within Dr. Rodrigo Gutierrez Laboratory from the Microbiology and Molecular Genetics Department of the Pontificia Universidad Católica.The Laboratory was searching to develop a software for data visualization.
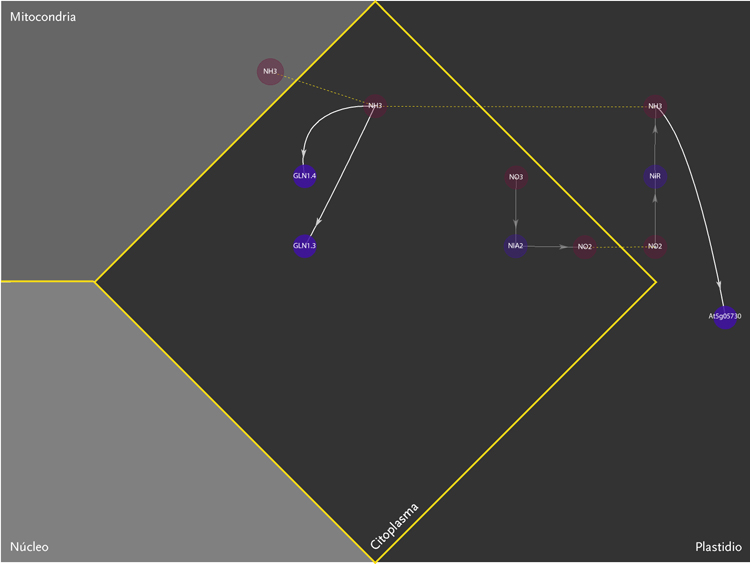
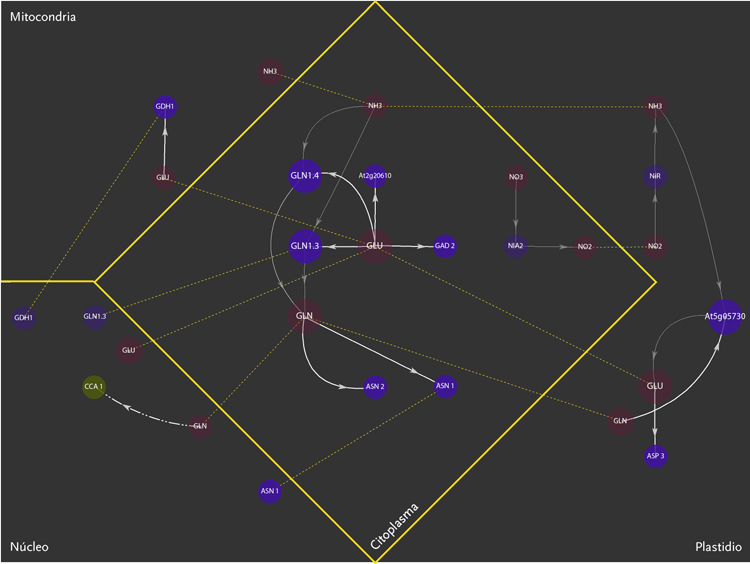
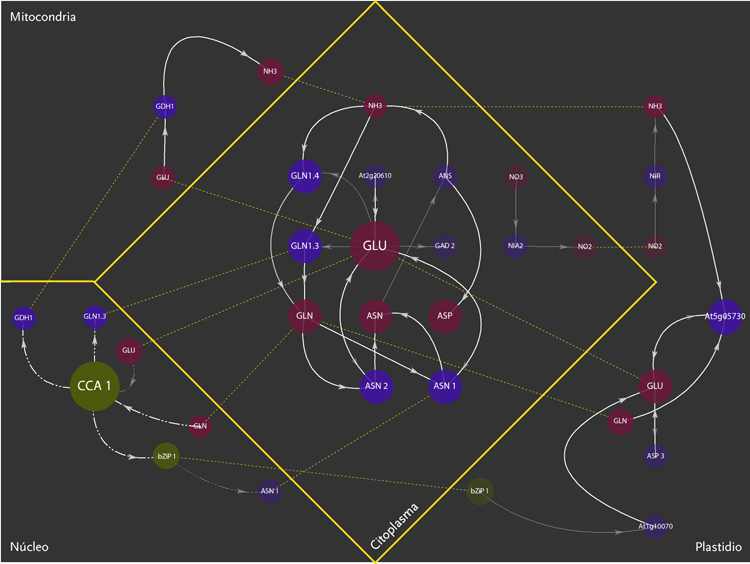
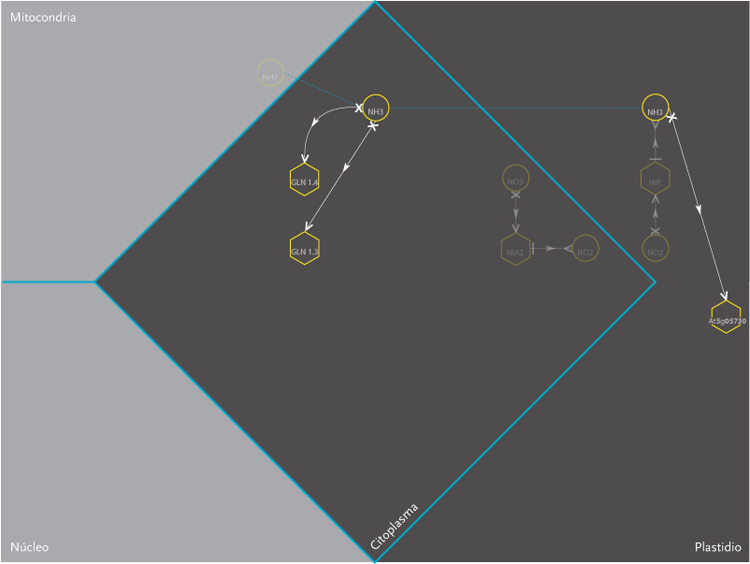
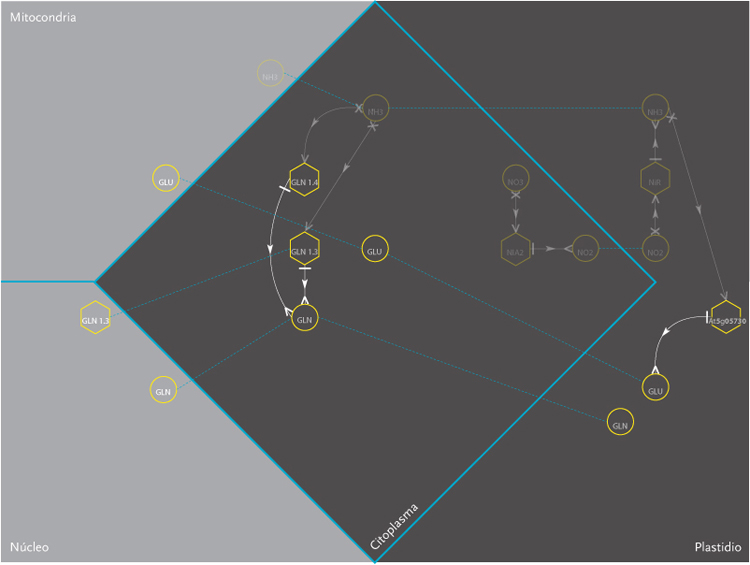
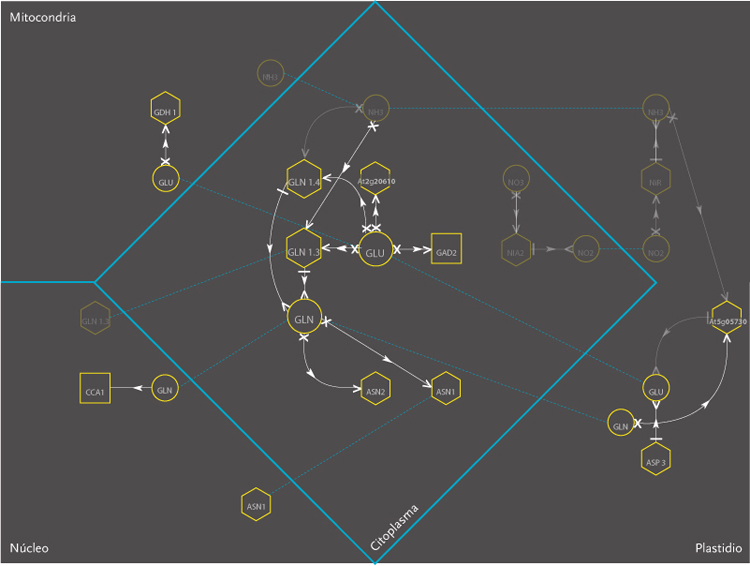
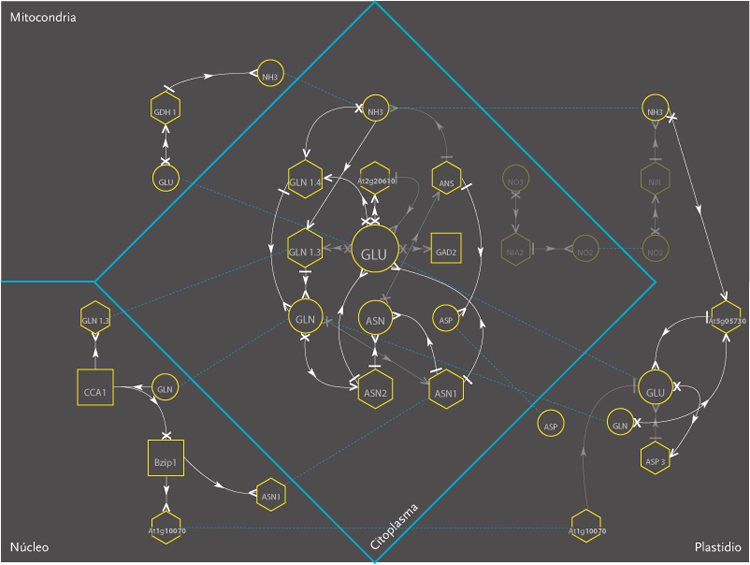
The purpose of the project was the design of a display model that could allow the observation of molecular interactions taking place inside a plant – Arabidopsis Thaliana – in response to a certain external stimulus, e.g. augmenting levels of Nitrogen in the soil. The data is composed of a network of agents and interactions. Agents are different kinds of molecules; metabolites, gens, enzymes,mRNA.Interactions can be understood as the agent’s capacity to modify its neighbor agents. Interactions depend on the agents properties. Another type of information aggregated was the location of molecules inside cellular compartments.
Molecular interaction networks II
Molecular interaction networks II
Completion: 2008
These images correspond to the work conducted within Dr. Rodrigo Gutierrez Laboratory from the Microbiology and Molecular Genetics Department of the Pontificia Universidad Católica. This time improving the visualization done on Molecular Interactions I. In this occasion an overall review of what had been achieved was pursued, and new amount of information was added to the previous model.
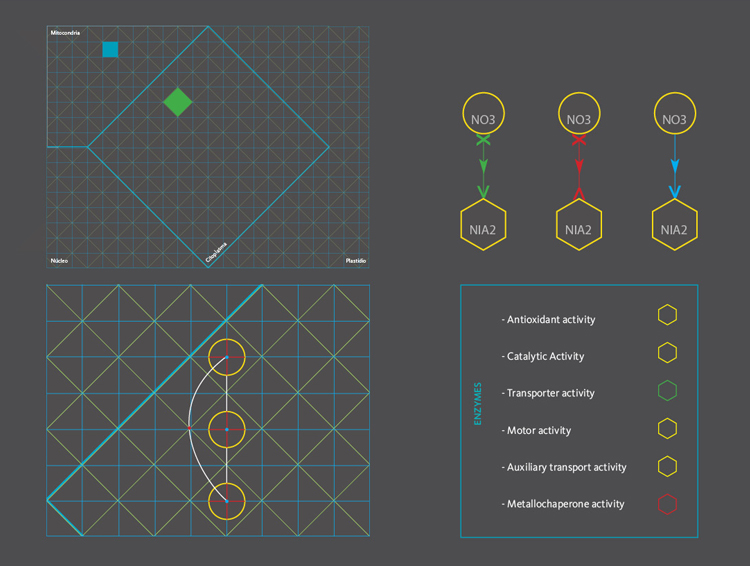
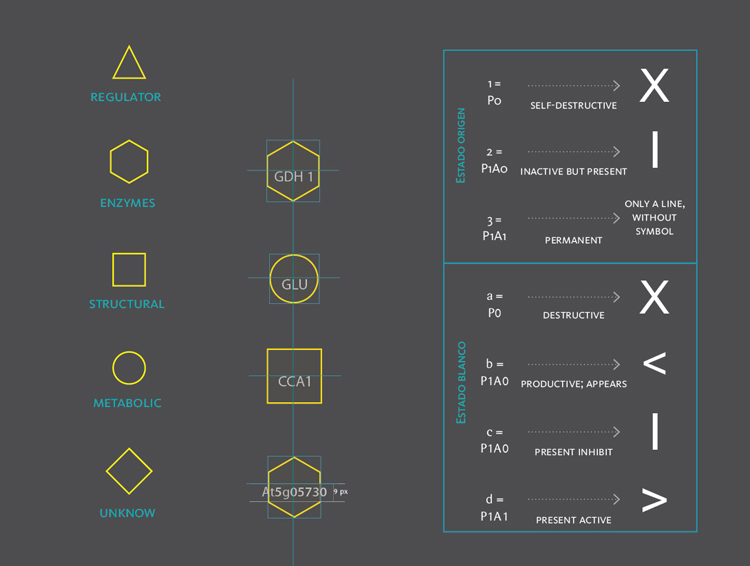
The new set of agents correspond to the same molecules of the former networks, yet this time divided in five different groups. These groups depend on molecular functions and had different geometric figures assigned. The types of interactions depend on the states of agents, which in turn, can be of two kinds; origin and target. A symbol belongs to each state. The combination of these symbols defines the interactions occurring within agents.The agents were divided in five groups and each one of them had specific belonging molecules. Per default, the agents of the all five groups were coloured yellow. Nevertheless the user had the option to change the colour of either the molecules or the complete group and one or more kind of interactions.